Semi-automatic segmentation by propagation
Computational anatomy aims to develop computer processing methods to enrich diagnosis by extracting objective and clinically useful information from medical images. The deployment of such methods remains limited for the exploration of soft tissue organs, especially for the study of pelvic floor disorders and neuromuscular diseases via magnetic resonance imaging (MRI). In these domains, the segmentation step is essential to allow the characterization of physiological alterations occurring within the organs of interest. The high phenotypic variability in these pathologies has so far limited the development of robust automatic segmentation methods, limiting clinical research on large populations. To overcome this challenge, the main contribution of my thesis was the development of a supervised segmentation approach based on diffeomorphic image registration propagation methods. The objective of this approach was to simplify the segmentation of image series presenting a continuity of information between successive images, whether they are time series or three-dimensional images.
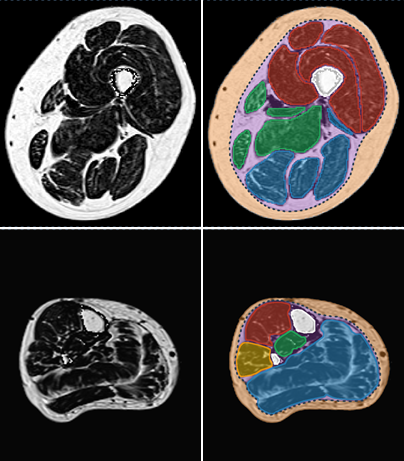
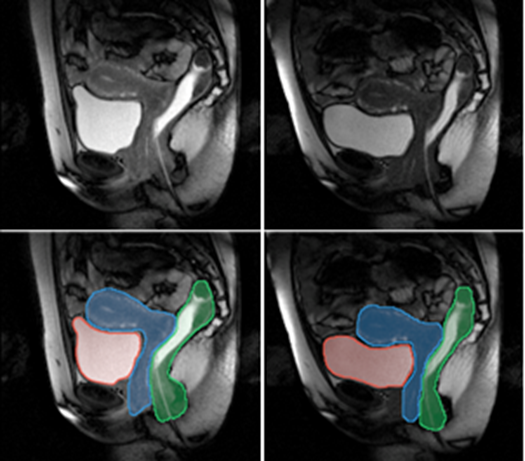
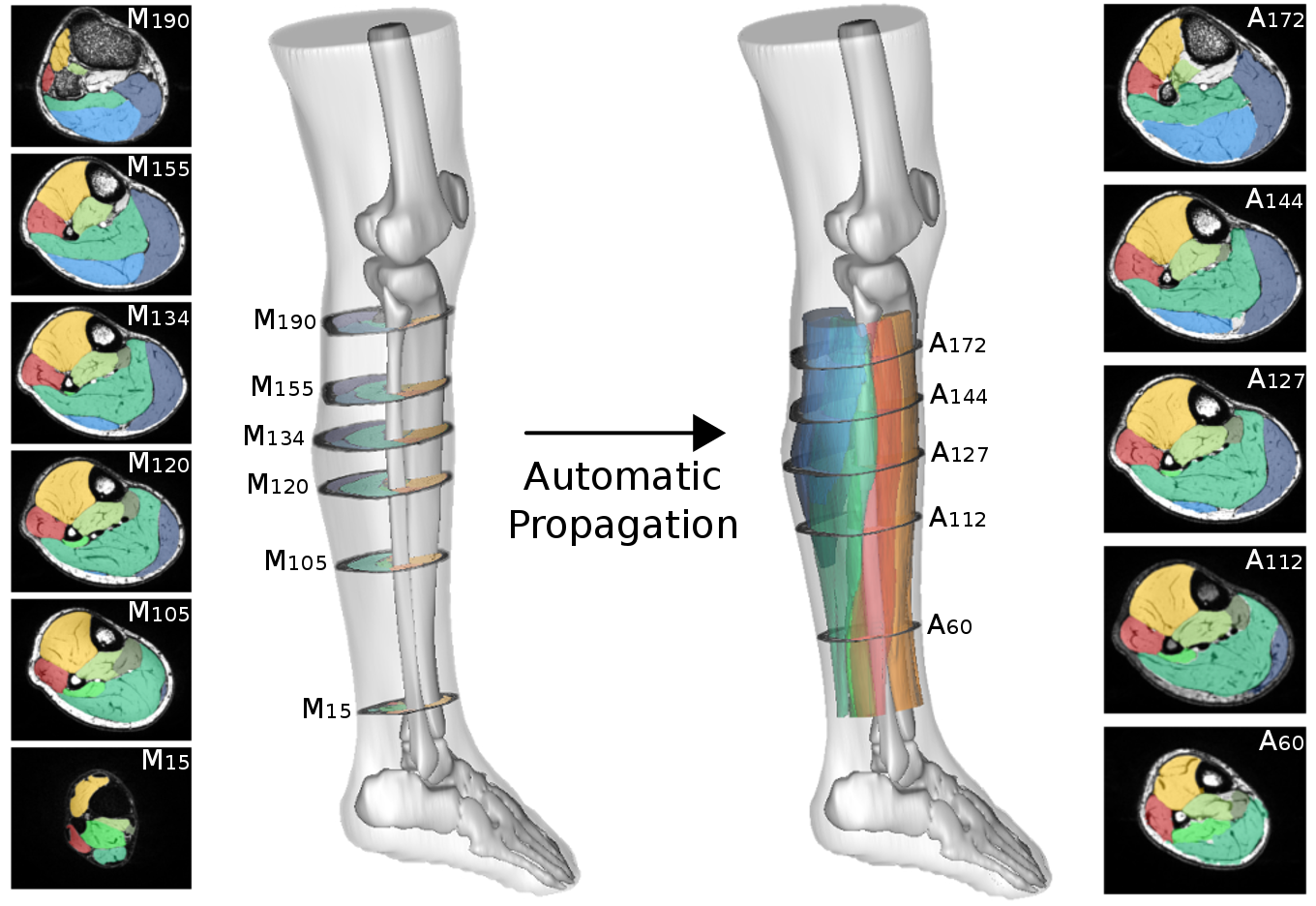
The method we have proposed exploits the redundancy of information contained between each successive slice of the image series acquired in our application contexts. Whether it is temporal image series for the pelvis or 3D spatial image series for the lower limbs, our data present a strong continuity from one slice to another. Thus, we proposed a semi-automatic method requiring only minimal manual segmentations from a user as a priori knowledge on the dataset in order to propagate the information contained in this manual input to the whole dataset. More concretely, since our image series are composed of several successive 2D slices, the objective was to manually provide segmentations of the regions of interest on a limited number of slices in order to propagate these segmentations to all other slices of the image series. Contrary to atlas-based or learning-based methods requiring the prior complete segmentation of a large database of appropriate images (not necessarily available), our process allows any user to obtain the direct segmentation of a series of images by limiting the time dedicated to manual segmentation. Please refer to Ogier et al. 2017 for the complete algorithm and parametric details of our proposed propagation method.
By considerably reducing the manual involvement of an operator (about 80%) and by providing a robust and accurate result, this method has been validated for the segmentation of skeletal muscles as well as the bladder in pathological contexts. Our method is intended to be applicable to the segmentation of any soft tissue. Therefore, it has also been used for segmentation of epicardial muscle and fat tissue on cardiac cine MRI sequences.

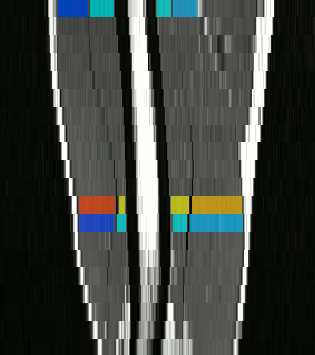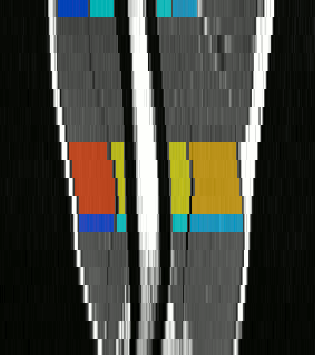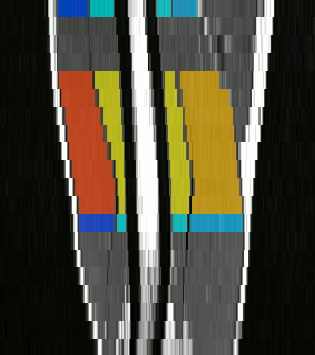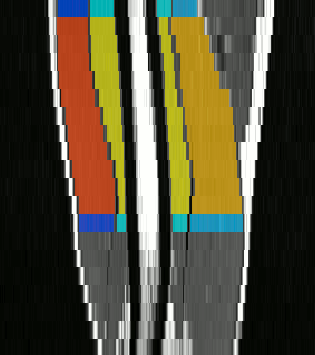
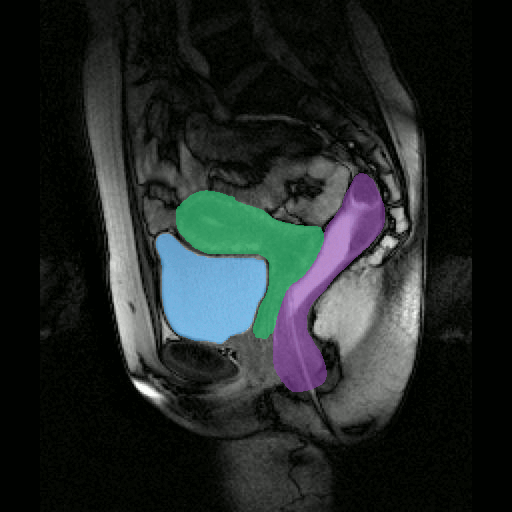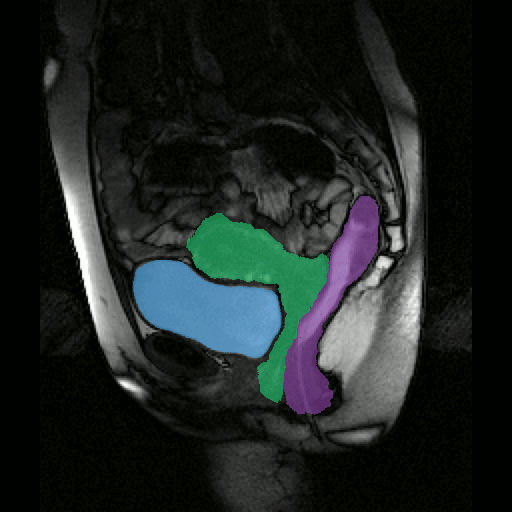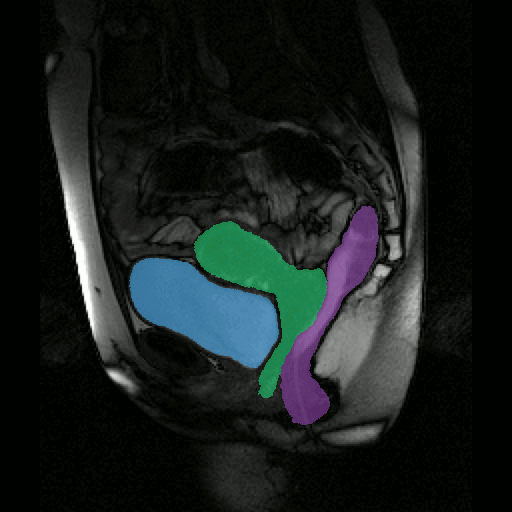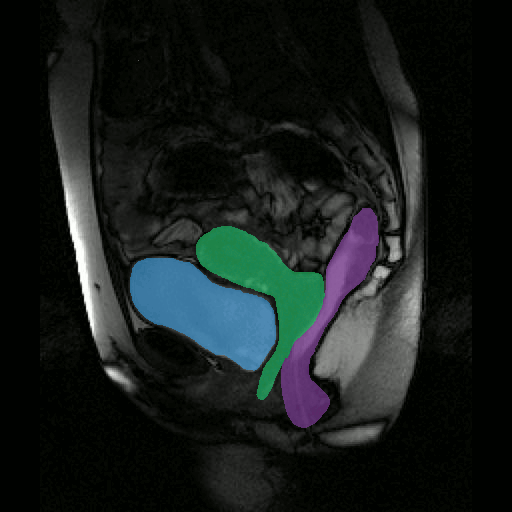
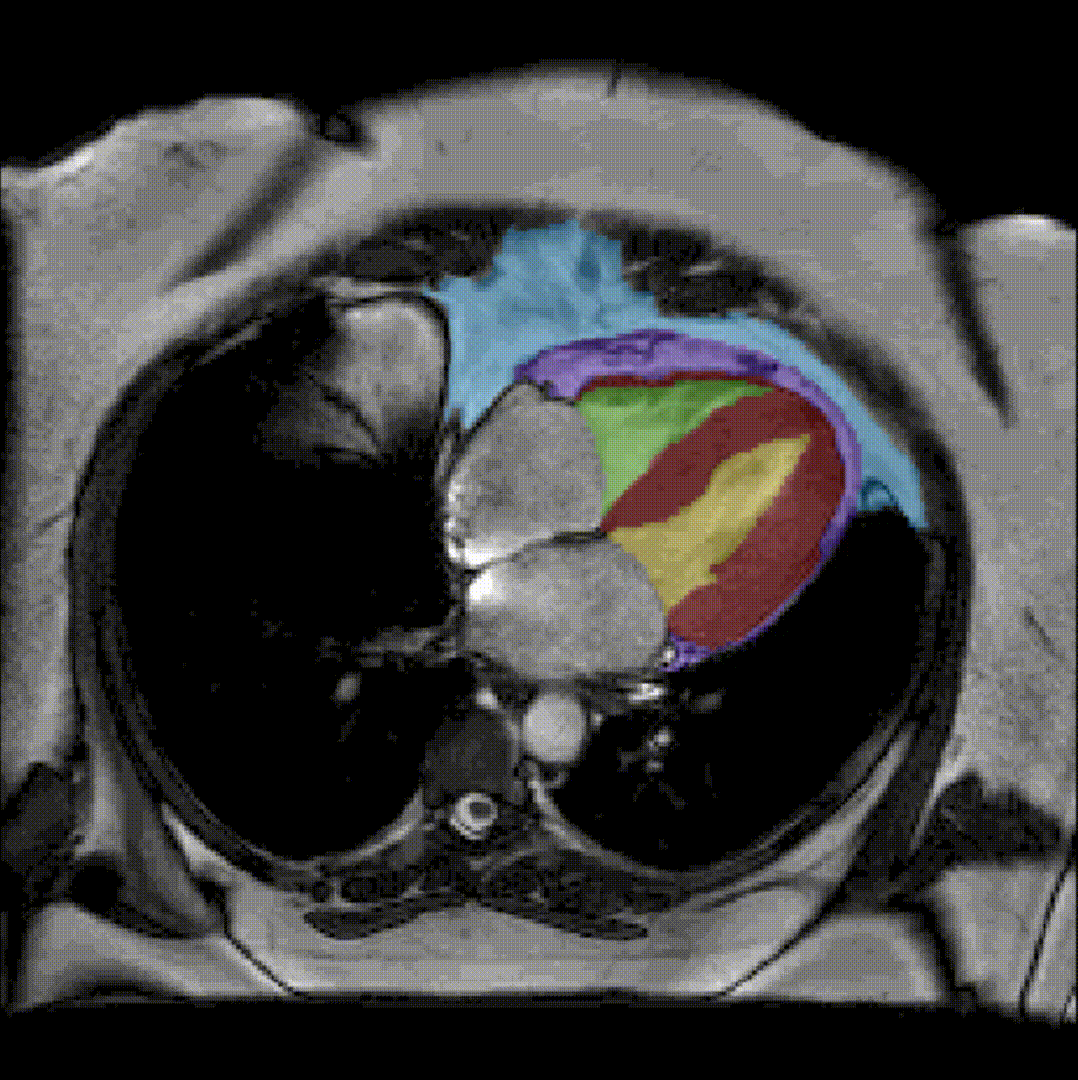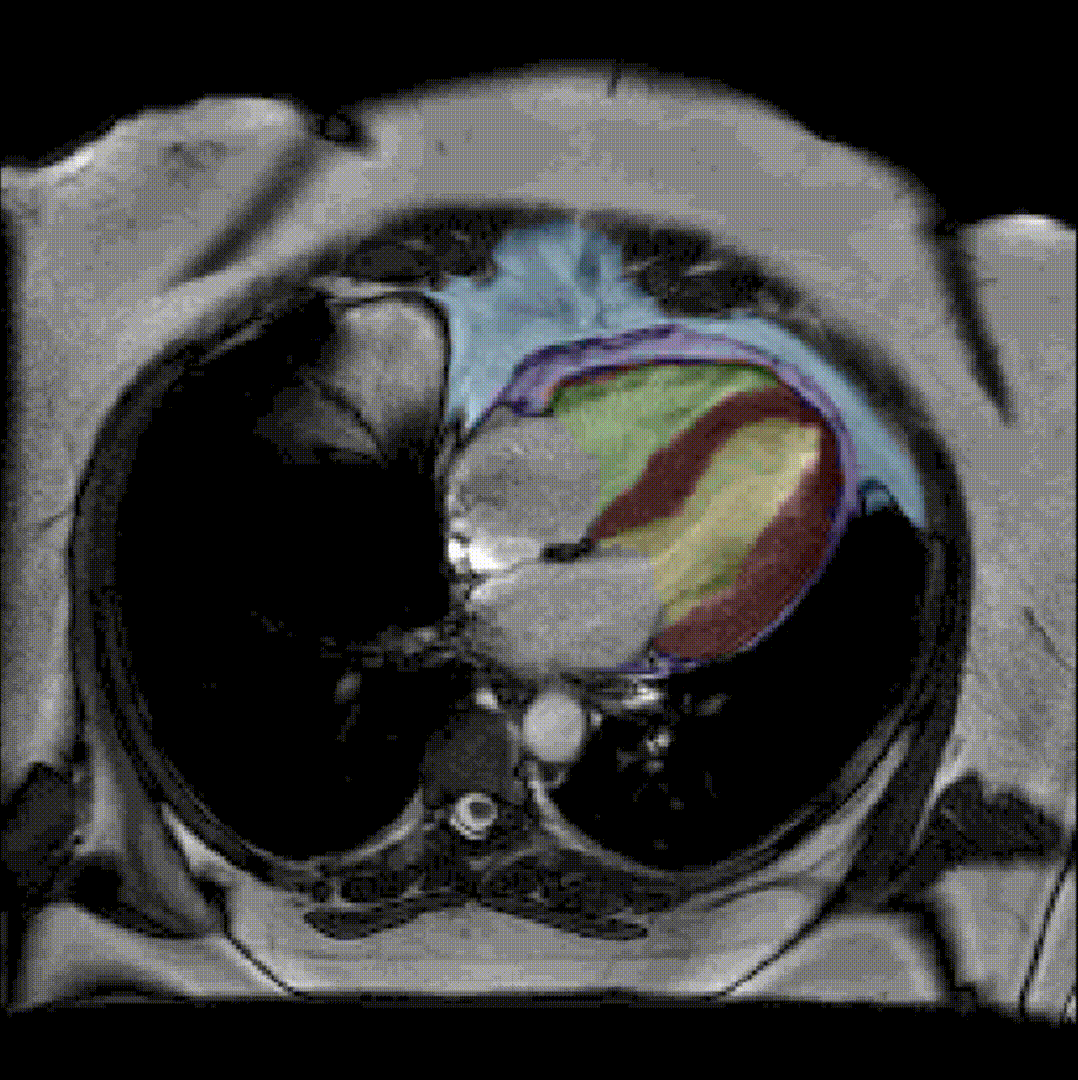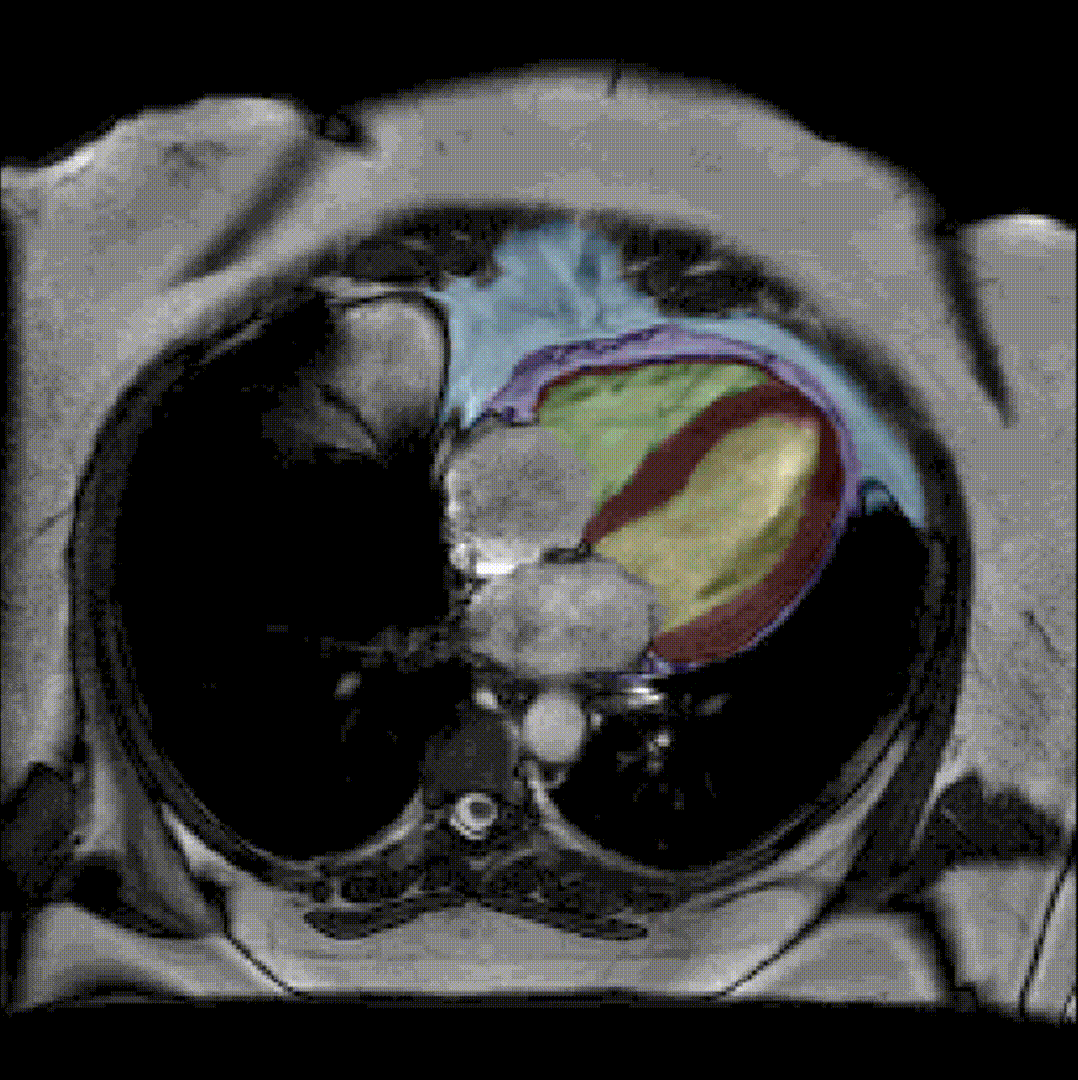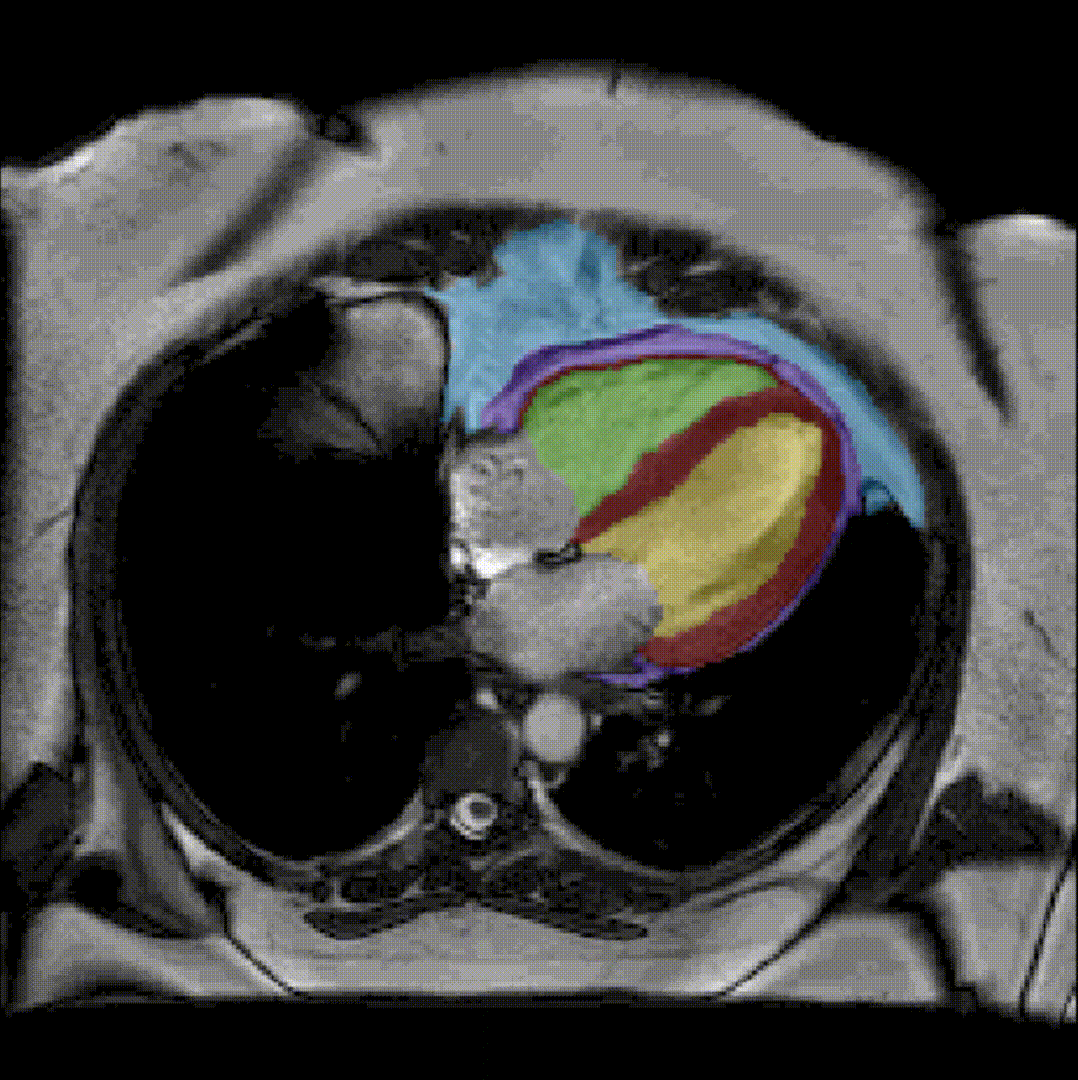
To further facilitate the study of neuromuscular diseases, our segmentation method has also been extended for the longitudinal follow-up of patients.
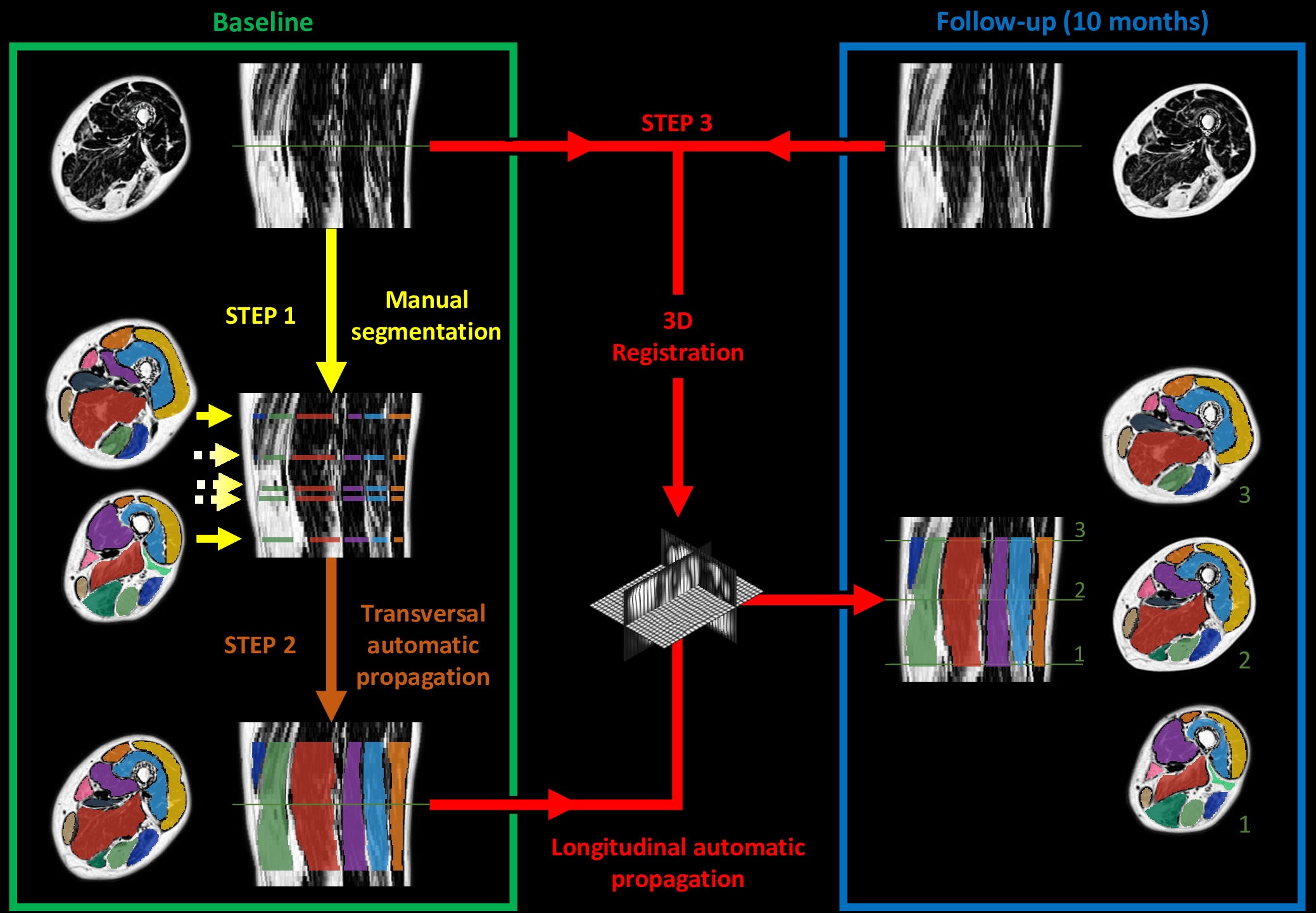
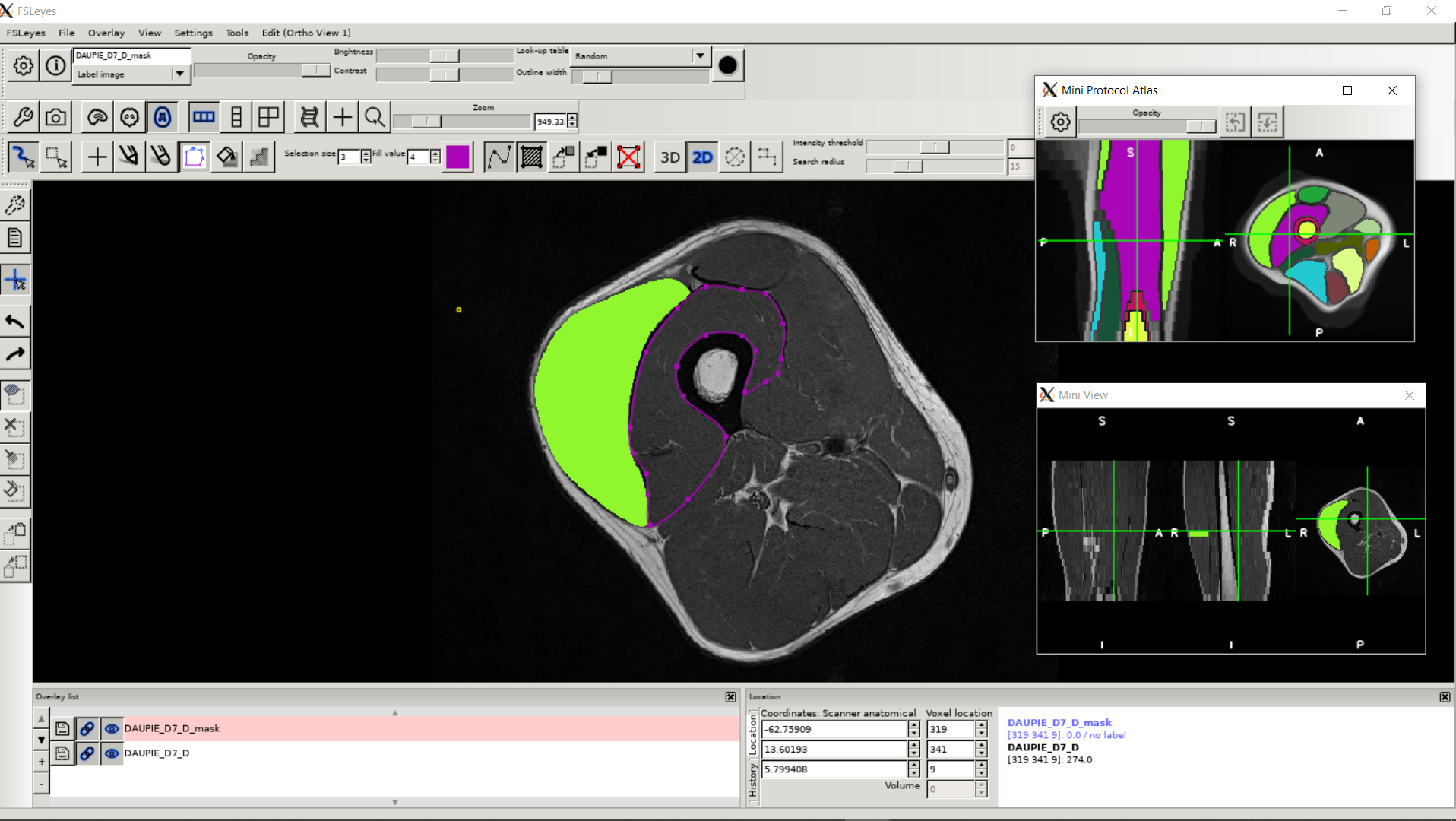
To date, no automatic approach has been proposed to facilitate the exploration of lower limbs MR images, to the point of limiting clinical research on large populations. Our proposed methods of segmentation have been directly applied in several clinical studies in order to characterize different neuromuscular diseases via the extraction of scores from quantitative MRI. See the dedicated page for these research projects. The segmentation protocol associated with our methodology has also been shown to be usable by a user who is not necessarily an expert in the field of image segmentation. A dedicated plugin has been implemented in the FSLeyes software to facilitate the distribution of our approaches to all researchers/clinicians.
Bibliography
- Patent
-
METHOD AND DEVICE FOR SEGMENTING IMAGES BY AUTOMATIC PROPAGATION INTO AN (N+1)-TH DIMENSION OF AN IMAGE SEGMENTATION INITIALIZED IN DIMENSION N
Arnaud Le Troter, David Bendahan, Augustin C. Ogier
Patent n°WO/2019/012220, 2019, France
- Journal articles
-
Overview of MR Image Segmentation Strategies in Neuromuscular Disorders
Augustin C. Ogier, Marc-Adrien Hostin, Marc-Emmanuel Bellemare, David Bendahan
Frontiers in Neurology, Frontiers, 2021, 12, pp.625308
DOI: 10.3389/fneur.2021.625308

-
Supervised segmentation framework for evaluation of diffusion tensor imaging indices in skeletal muscle
Laura Secondulfo, Augustin C. Ogier, Jithsa Monte, Vincent Aengevaeren, David Bendahan, Aart Nederveen, Gustav Strijkers, Melissa Hooijmans
NMR in Biomedicine, Wiley, 2020
DOI: 10.1002/nbm.4406

-
A novel segmentation framework dedicated to the follow‐up of fat infiltration in individual muscles of patients with neuromuscular disorders
Augustin C. Ogier, Linda Heskamp, Constance Michel, Alexandre Fouré, Marc-Emmanuel Bellemare, Arnaud Troter, Arend Heerschap, David Bendahan
Magnetic Resonance in Medicine, Wiley, 2019, 83, pp.pp.1825-1836
DOI: 10.1002/mrm.28030

- Conference papers
-
Individual muscle segmentation in MR images: A 3D propagation through 2D non-linear registration approaches
Augustin C. Ogier, Michaël Sdika, Alexandre Fouré, Arnaud Le Troter, David Bendahan
The 39th Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBC’17), Jul 2017, Seogwipo, South Korea, pp.317-320
DOI: 10.1109/EMBC.2017.8036826

- Conferencecommunications
-
Méthode de segmentation dédiée au suivi de l'infiltration en graisse des muscles individuels des patients atteints de maladies neuromusculaires
Augustin C. Ogier, Linda Heskamp, Alexandre Fouré, Arnaud Le Troter, Arend Heerschap, David Bendahan
16es Journées de la Société Française de Myologie (JSFM), Nov 2018, Brest, France
-
Semi-automatic segmentation of individual muscles in MR images: A new tool dedicated to the follow-up of patients with neuromuscular disorders
Augustin C. Ogier, Linda Heskamp, Alexandre Fouré, Marc-Emmanuel Bellemare, Arnaud Le Troter, Arend Heerschap, David Bendahan
International Society for Magnetic Resonance in Medicine (ISMRM), Jun 2018, Paris, France
-
An accurate supervised segmentation of 3D individual lower leg muscles from 7T-MRI using label propagation
Augustin C. Ogier, Alexandre Fouré, Michaël Sdika, Arnaud Le Troter, David Bendahan
MYO-MRI, Imaging in Neuromuscular Disease, Nov 2017, Berlin, Germany
-
3D MRI Segmentation of muscle through 2D multi-label propagation at 7T
Augustin C. Ogier, Alexandre Fouré, Michaël Sdika, Arnaud Le Troter, David Bendahan
The 34th Annual Scientific Meeting - European Society for Magnetic Resonance in Medicine and Biology (ESMRMB), Oct 2017, Barcelone, Spain
-
Segmentation des muscles individuels en IRM basée sur des approches de recalage non-linéaire
Augustin C. Ogier, Michaël Sdika, Alexandre Fouré, Arnaud Le Troter, David Bendahan
3ème Congrès de la Société Française de Résonance Magnétique en Biologie et Médecine (SFRMBM), Mar 2017, Bordeaux, France
-
Segmentation d’images IRM à 7T des muscles individuels basée sur des approches de propagation de masques
Augustin C. Ogier, Alexandre Fouré, Michaël Sdika, Arnaud Le Troter, David Bendahan
15es Journées de la Société Française de Myologie (JSFM), Nov 2017, Colmar, France
